-
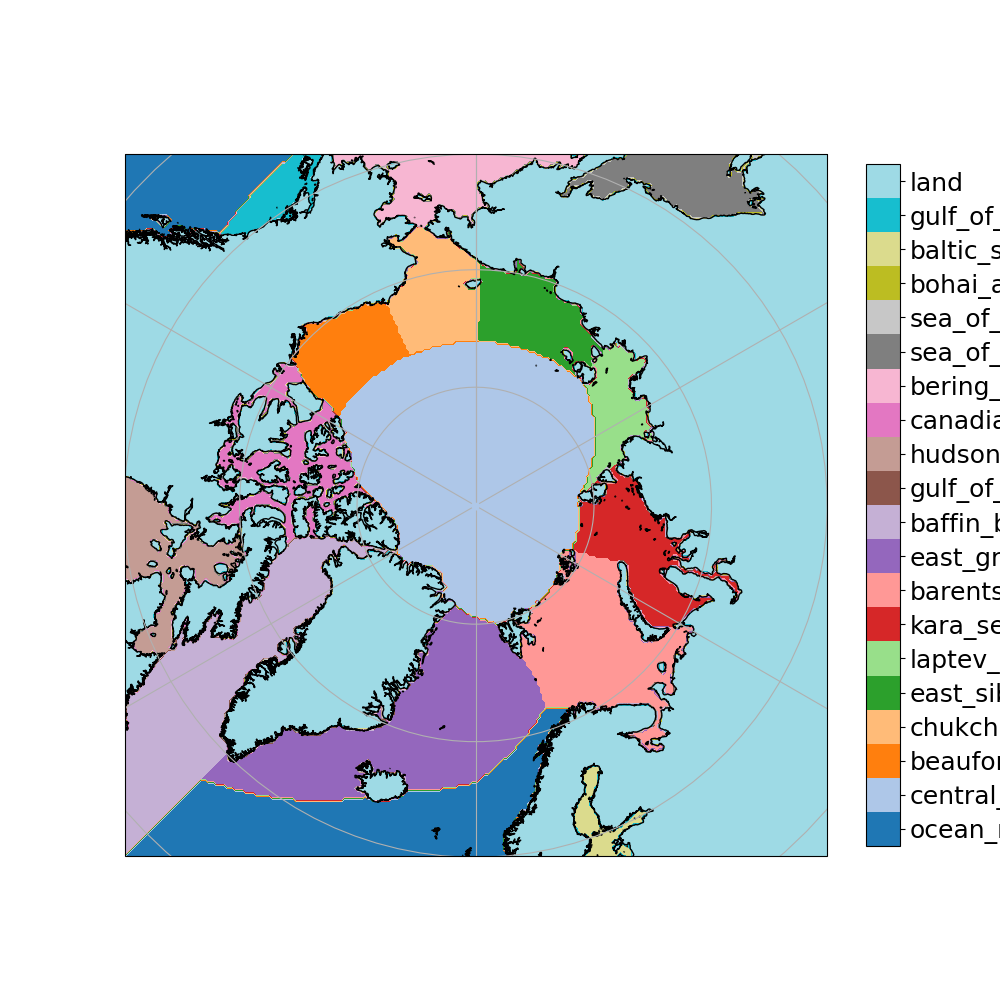
This is the netCDF file which is used for the regional masks: NSIDC-0780_SeaIceRegions_EASE2-N25km_v1.0.nc and this is the one with the land/ice/ocean mask: EASE2_N25km.LOCImask_land50_coast0km.720x720.bin
Edited by Valentin Ludwig
Please sign in to comment